
BioExcel Webinar: A Virtual Research Environment for Integrative Modelling of Biomolecular Complexes with the New Modular Version of HADDOCK
BioExcel, the leading European Centre of Excellence for computational biomolecular research, aims to support Life Science academic and industrial researchers in the effective use of HPC biomolecular software and continues BioExcel’s webinar series with its top experts in biomolecular modelling and simulations tools. Welcome!
Webinar: A Virtual Research Environment for Integrative Modelling of Biomolecular Complexes with the New Modular Version of HADDOCK
| Date: | June 13, 2023 |
| Time: | 15:00 CEST (Central European Summer Time) UTC/GMT + 2h |
| 16:00 EEST (Eastern European Summer Time) UTC/GMT + 3 h | |
| Please do not forget to take into consideration the different time zones of joining the webinar! | |
| Duration: | 1 hour |
| Tool: | ZOOM (install the latest Zoom application before the webinar via https://zoom.us/download) |
Abstract
The prediction of the quaternary structure of biomolecular macromolecules is of paramount importance for fundamental understanding of cellular processes and drug design. In the era of integrative structural biology, one way of increasing the accuracy of modelling methods used to predict the structure of biomolecular complexes is to include as much experimental or predictive information as possible in the process. We have developed for this purpose a versatile information-driven docking approach HADDOCK (https://www.bonvinlab.org/software) available as a web service at https://wenmr.science.uu.nl/haddock2.4. HADDOCK can integrate information derived from biochemical, biophysical or bioinformatics methods to guide the modelling.
In the context of the BioExcel Center of Excellence for Computational Biomolecular Research (https://bioexcel.eu), we have developed HADDOCK3, the new modular version of HADDOCK. It represents a redesign of the HADDOCK2.X series, implementing new ways to interact with the HADDOCK sub-routines and offering more customization. Users can create custom workflows by combining different modules, thus making the workflows tailored to their specific needs. HADDOCK3 has therefore developed to truthfully work like a puzzle of many pieces (simulation modules) that users can combine to more accurately model their systems. The HADDOCK3 workflows are defined in straightforward configuration files, similar to the TOML format (also supported).
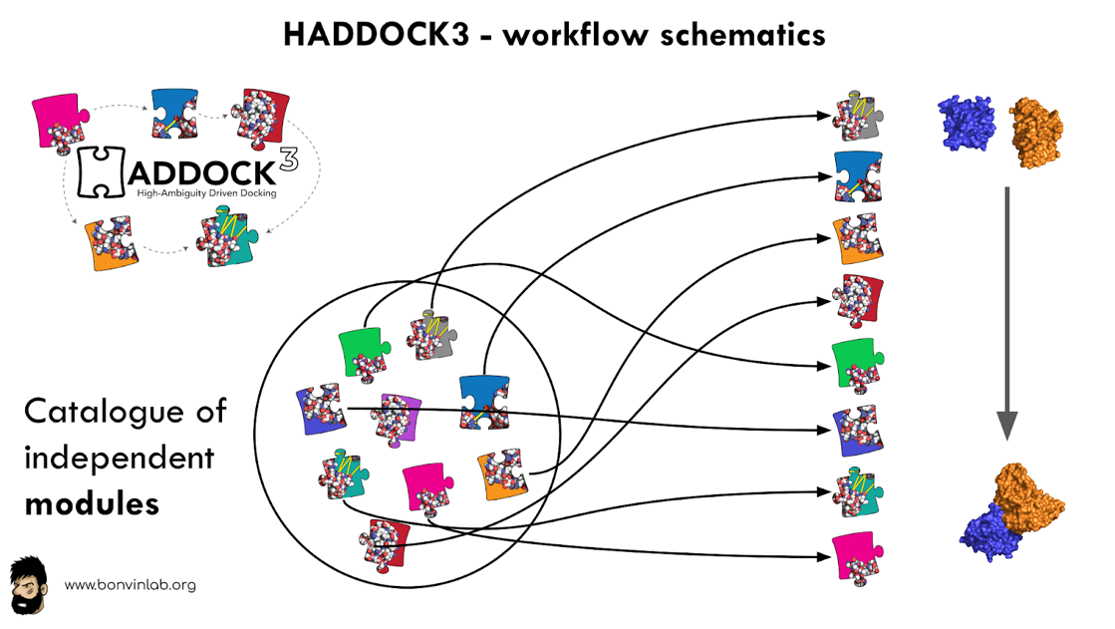
In order to facilitate the use of HADDOCK3, in collaboration with the Netherlands eScience Center (https://www.esciencecenter.nl) we are developing a customizable, interactive, HTC/Cloud (and HPC)-optimized and reusable Virtual Research Environment (VRE) for Integrative Modelling of Biomolecular Complexes (https://github.com/i-VRESSE). By integrating all steps involved in studying biomolecular interactions, this VRE will lower the steep learning curve for researchers and students from different fields and contribute to reproducible research and FAIR sharing of data.
In this webinar, I will introduce HADDOCK3 and discuss the status of the Virtual Research Environment for Integrative Modelling of Biomolecular Complexes.
Presenters
- Alexandre Bonvin (Professor of Computational Structural Biology)
More information on the presenter in the presenters tab!
Registration
Please register below! You will then receive a confirmation email with details of how you can connect to the webinar.
Time
13.6.2023 16:00 - 17:00